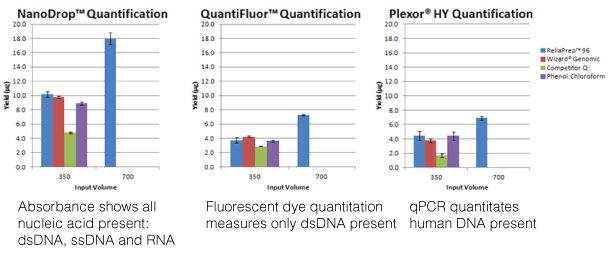
With the advent of genome editing using CRISPR-Cas9, researchers have been excited by the possibilities of precisely placed edits in cellular DNA. Any double-stranded break in DNA, like that induced by CRISPR-Cas9, is repaired by one of two pathways: Non-homologous end joining (NHEJ) or homology-directed repair (HDR). Using the NHEJ pathway results in short insertions or deletions (indels) at the break site, so the HDR pathway is preferred. However, the low efficiency of HDR recombination to insert exogenous sequences into the genome hampers its use. There have been many attempts at boosting HDR frequency, but the methods compromise cell growth and behave differently when used with various cell types and gene targets. The strategy employed by the authors of an article in Communications Biology tethered the DNA donor template to Cas9 complexed with the ribonucleoprotein and guide RNA, increasing the local concentration of the donor template at the break site and enhancing homology-directed repair. Continue reading “All You Need is a Tether: Improving Repair Efficiency for CRISPR-Cas9 Gene Editing”