Now that Promega is expanding its offerings of options for examining live-cell protein interactions or quantitation at endogenous protein expression levels, we in Technical Services are getting the question about which option is better. The answer is, as with many assays… it depends! First let’s talk about what are the NanoBiT and NanoBRET technologies, and then we will provide some similarities and differences to help you choose the assay that best suits your individual needs.
Continue reading “A BiT or BRET, Which is Better?”protein:protein interactions
Studying Mitochondrial Fission with NanoBiT Complementation Assay
Updated February 15, 2021
If you’re interrogating two proteins to understand the conditions under which they interact, a complementation system enables you to tag each protein. Interaction of the tagged proteins facilitates the complementation of the subunits, resulting in a signal. Here we discuss the NanoBiT complementation assay and describe its use to study mitochondrial fission.
Continue reading “Studying Mitochondrial Fission with NanoBiT Complementation Assay”NanoBiT™ Assay: Transformational Technology for Studying Protein Interactions Named a Top 10 Innovation of 2015

For three out of the last four years, we have been honored to have one of our key technologies named a Top 10 Innovation by The Scientist. This year the innovative NanoBiT™ Assay (NanoLuc® Binary Technology) received the recognition. NanoBiT™ is a structural complementation reporter based on NanoLuc® Luciferase, a small, bright luciferase derived from the deep sea shrimp Oplophorus gracilirostris.
Using plasmids that encode the NanoBiT complementation reporter, you can make fusion proteins to “report” on protein interactions that you are studying. One of the target proteins is fused to the 18kDa subunit; the other to the 11 amino acid subunit. The NanoBiT™ subunits are stable, exhibiting low self-affinity, but produce an ultra-bright signal upon association. So, if your target proteins interact, the two subunits are brought close enough to each other to associate and produce a luminescent signal. The strong signal and low background associated with a luminescent system, and the small size of the complementation reporter, all help the NanoBiT™ assay overcome the limitations associated with traditional methods for studying protein interactions.
The small size reduces the chances of steric interference with protein interactions. The ultra bright signal, means that even interactions among proteins present in very low amounts can be detected and quantified–without over-expressing large quantities of non-native fusion proteins and potentially disrupting the normal cellular environment. And the NanoBiT™ assay can be performed in real time, in live cells.
The NanoBiT™ assay is already being deployed in laboratories to help advance understanding of fundamental cell biology. You can see how one researcher is already taking full advantage of this innovative technology in the video embedded below:
Visit the Promega web site to see more examples more examples how the NanoBiT™ assay can break through the traditional limitations for studying protein interactions in cells.
You can read the Top 10 article in The Scientist here.
About the Development of an Improved BRET Assay: NanoBRET
One of the more exciting reporter molecules technologies available came online in the past year, with the launch of the Promega NanoBRET™ technology. While it’s easy for me, a science writer at Promega, to brag, seriously, this is a very cool protein interactions tool.
A few of the challenges facing protein-protein interactions researchers include:
- The ability to quantitatively characterize protein-protein interactions
- Ability to examine protein-protein interactions in situ, in the context of the living cell
A goal of the NanoBRET™ developers was to improve the sensitivity and dynamic range of traditional BRET technology, in order to address these challenges.
In May 2015 these researchers published an article outlining their efforts to create NanoBRET technology in ACS Chemical Biology, in an article entitled, “NanoBRET—A Novel BRET Platform for the Analysis of Protein-Protein Interactions”. Here is a brief look at their work.
Continue reading “About the Development of an Improved BRET Assay: NanoBRET”
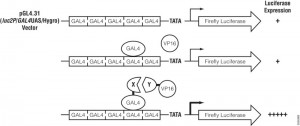
CheckMate™ Mammalian Two-Hybrid System: Application Update
In the CheckMate™ Mammalian Two-Hybrid System, the pBIND Vector contains the yeast GAL4 DNA-binding domain upstream of a multiple cloning region, and the pACT Vector contains the herpes simplex virus VP16 activation domain upstream of a multiple cloning region. The two genes encoding the two potentially interactive proteins of interest are cloned into pBIND and pACT Vectors to generate fusion proteins with the DNA-binding domain of GAL4 and the activation domain of VP16, respectively. The pG5luc Vector contains five GAL4 binding sites upstream of a minimal TATA box, which in turn, is upstream of the firefly luciferase gene (luc+). The pGAL4 and pVP16 fusion constructs are transfected along with pG5luc Vector into mammalian cells. Interaction between the two test proteins, as GAL4 and VP16 fusion constructs, results in an increase in firefly luciferase expression over the negative controls. Traditionally mammalian two hybrid analysis was used to confirm initial data obtained from yeast two hybrid experiments.
Due to enhanced bioinformatics information and the development of improved co-immunoprecipiation/pulldown procedures/technology, there is a growing trend to use only mammalian cells to characterize protein:protein interactions. The following references illustrate the use of the CheckMate™ system to complement other techniques to characterize protein;protein interactions using only mammalian cells .
Bagchi, P. et al. (2013) Molecular Mechanism behind Rotavirus Nsp-1 Mediated PI3 Kinase Activation: Interaction between NSP1 and the p85Subunit of PI3 kinase. J. Vir. 87, 2358-62.
Greninger, A. et al. (2013) ACBD3 Interaction with TBC1 domain 22 protein is differentially affected by Enteroviral and Kobuviral 3A protein binding. mBio 4, 00098-13.
Patki, M. et al. ( 2013) The ETS Domain Transcription Factor ELK1 Direct a critical component of growth signalling by Androgen Receptor in prostrate cancer cells. J.Biol. Chem. 288 11047-65
Konig, H-G. (2012) Fibroblast growth factor homologous factor 1 interact with NEMO to regulate NF-kappaB signaling in neurons J. Cell Sci. 125, 6058-70
Protein:protein Interactions: GST Pulldowns
 GST pull-downs
GST pull-downs
Pull-down assays probe interactions between a protein of interest that is expressed as a fusion protein (e.g., bait) and the potential interacting partners (prey). In a pull-down assay one protein partner is expressed as a fusion protein (e.g., bait protein) in E. coli and then immobilized using an affinity ligand specific for the fusion tag. The immobilized bait protein can then be incubated with the prey protein. The source of the prey protein can be either from a cell-based or cell-free expression system. After a series of wash steps the entire complex can be eluted from the affinity support using SDS-PAGE loading buffer or by competitive analyte elution, then evaluated by SDS-PAGE.
Continue reading “Protein:protein Interactions: GST Pulldowns”