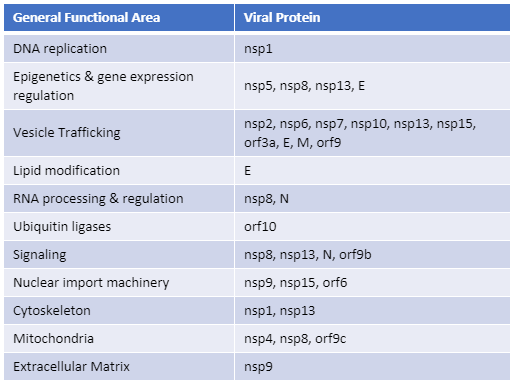
For three out of the last four years, we have been honored to have one of our key technologies named a Top 10 Innovation by The Scientist. This year the innovative NanoBiT™ Assay (NanoLuc® Binary Technology) received the recognition. NanoBiT™ is a structural complementation reporter based on NanoLuc® Luciferase, a small, bright luciferase derived from the deep sea shrimp Oplophorus gracilirostris.
Using plasmids that encode the NanoBiT complementation reporter, you can make fusion proteins to “report” on protein interactions that you are studying. One of the target proteins is fused to the 18kDa subunit; the other to the 11 amino acid subunit. The NanoBiT™ subunits are stable, exhibiting low self-affinity, but produce an ultra-bright signal upon association. So, if your target proteins interact, the two subunits are brought close enough to each other to associate and produce a luminescent signal. The strong signal and low background associated with a luminescent system, and the small size of the complementation reporter, all help the NanoBiT™ assay overcome the limitations associated with traditional methods for studying protein interactions.
The small size reduces the chances of steric interference with protein interactions. The ultra bright signal, means that even interactions among proteins present in very low amounts can be detected and quantified–without over-expressing large quantities of non-native fusion proteins and potentially disrupting the normal cellular environment. And the NanoBiT™ assay can be performed in real time, in live cells.
The NanoBiT™ assay is already being deployed in laboratories to help advance understanding of fundamental cell biology. You can see how one researcher is already taking full advantage of this innovative technology in the video embedded below:
Visit the Promega web site to see more examples more examples how the NanoBiT™ assay can break through the traditional limitations for studying protein interactions in cells.
You can read the Top 10 article in The Scientist here.
Like this:
Like Loading...