You have identified and cloned your protein of interest, but you want to explore its function. A protein fusion tag might help with your investigation. However, choosing a tag for your protein depends on what experiments you are planning. Do you want to purify the protein? Would you like to identify interacting proteins by performing pull-down assays? Are you interested in examining the endogenous biology of the protein? Here we cover the advantages and disadvantages of some protein tags to help you select the one that best suits your needs.
Affinity Tags
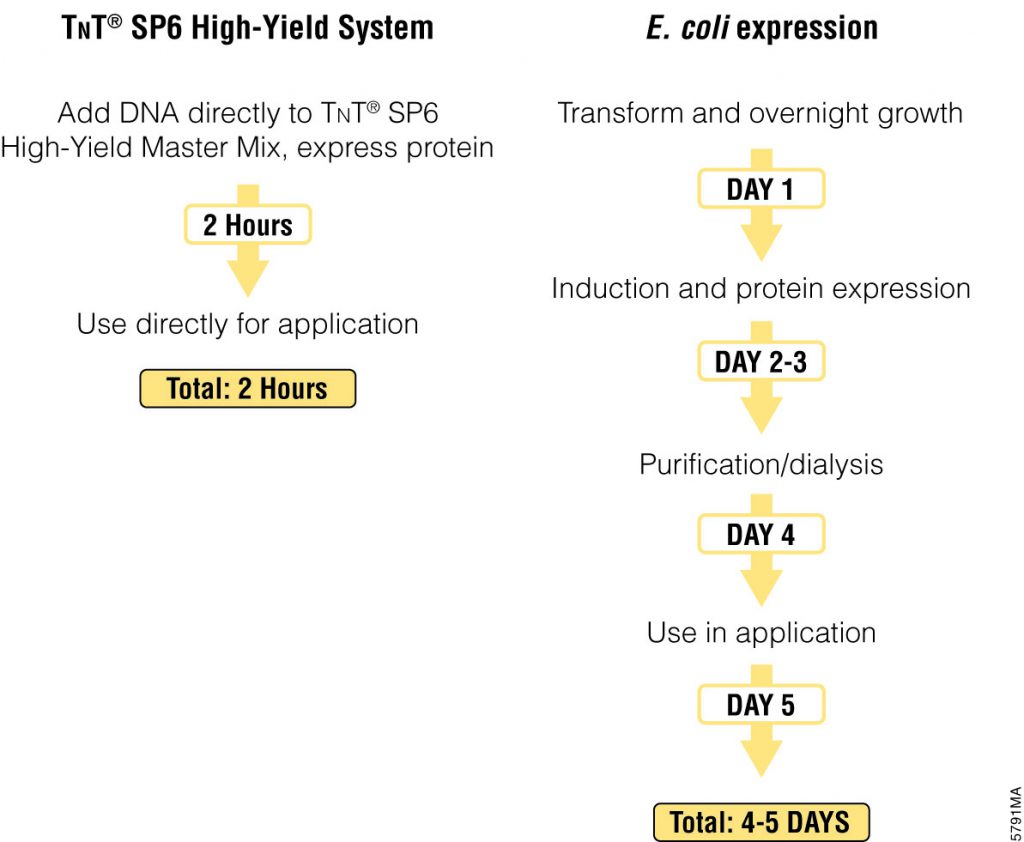
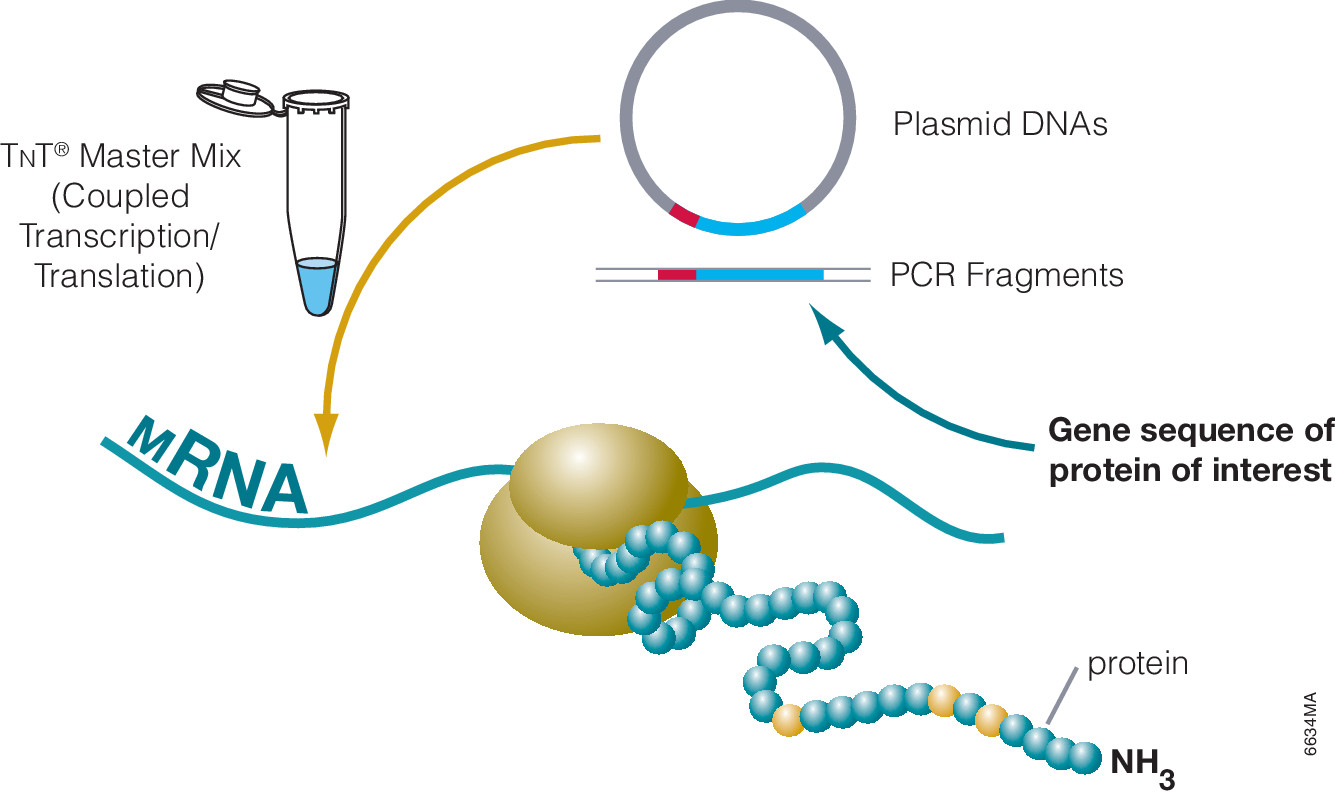
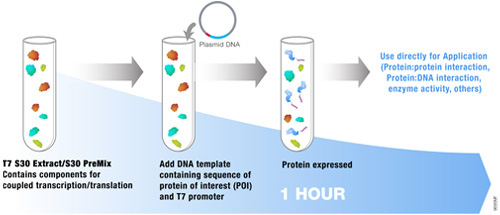
The most commonly used protein tags fall under the category of affinity tags. This means that the tag binds to another molecule or metal ion, making it easy to purify or pull down your protein of interest. In all cases, the tag will be fused to your protein of interest at either the amino (N) or carboxy (C) terminus by cloning into an expression vector. This protein fusion can then be expressed in cells or cell-free systems, depending on the promoter the vector contains.
Continue reading “Choosing a Tag for Your Protein”