Often a diagnosis of thyroid cancer is associated with a good prognosis and fairly straightforward surgical treatments to remove the tumor followed by radioactive iodine ablation. Such treatment works well in tumors that have not metastasized and retain enough of their thyroid cell “identity” that they can still accumulate radioactive iodine.
Often a diagnosis of thyroid cancer is associated with a good prognosis and fairly straightforward surgical treatments to remove the tumor followed by radioactive iodine ablation. Such treatment works well in tumors that have not metastasized and retain enough of their thyroid cell “identity” that they can still accumulate radioactive iodine.
However, aggressive thyroid cancers, which often metastasize and recur, do not respond to standard treatments because they are generally too dedifferentiated to accumulate iodine, so alternative treatments are needed.
One approach is to look for compounds that will reverse dedifferentiation, making tumor cells more likely to take up and concentrate radioactive iodine regardless of their location in the body. One possible target to effect dedifferentiation is epigenetic modification of histone proteins.
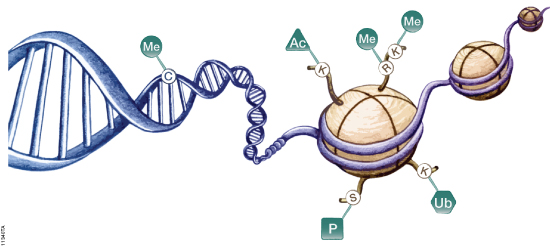
Histone proteins are more than the structural components of the nucleosome that organizes the chromatin inside cells. Histone proteins are subject to a host of protein modifications on their N-terminal tails such as acetylation, phosphorylation, methylation, ubiquitination and ADP-ribosylation. These various modifications are seen as creating a “histone code” that is read by other proteins and protein complexes (1). This code regulates patterns of gene expression and activity for a cell—in part resulting in a differentiated phenotype. Previous studies have suggested that some histone deacetylase (HDAC) inhibitors (e.g., valproic acid) can reverse some of the dedifferentiation associated with aggressive cancers (2).
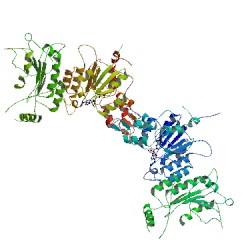
Jang, et al. in a recent paper (3) published in Cancer Gene Therapy synthesized a group of HDAC inhibitor analogs (AB1–AB13) and tested them for their ability to inhibit growth of three aggressive human thyroid cancer cell lines and induce partial re-differentiation to the thyroid cell phenotype. Continue reading “Rewriting the Histone Code: Searching for Treatments for Stage IV Thyroid Cancers”
Like this:
Like Loading...

 Often a diagnosis of thyroid cancer is associated with a good prognosis and fairly straightforward surgical treatments to remove the tumor followed by radioactive iodine ablation. Such treatment works well in tumors that have not metastasized and retain enough of their thyroid cell “identity” that they can still accumulate radioactive iodine.
Often a diagnosis of thyroid cancer is associated with a good prognosis and fairly straightforward surgical treatments to remove the tumor followed by radioactive iodine ablation. Such treatment works well in tumors that have not metastasized and retain enough of their thyroid cell “identity” that they can still accumulate radioactive iodine.

 Imagine that you are sitting in your room when you smell cherries and you are suddenly, inexplicably afraid. Although odors can elicit strong emotional responses, you have no bad memories of cherries. What you don’t know is that your father did, and you have inherited his fear. Sound far fetched? Maybe not. A paper published in Nature Neuroscience found just such an inherited association in mice (1).
Imagine that you are sitting in your room when you smell cherries and you are suddenly, inexplicably afraid. Although odors can elicit strong emotional responses, you have no bad memories of cherries. What you don’t know is that your father did, and you have inherited his fear. Sound far fetched? Maybe not. A paper published in Nature Neuroscience found just such an inherited association in mice (1).