Therapeutic monoclonal antibodies are large, complex molecules that undergo numerous post translational modifications (PTMs). In-depth characterization of antibody PTMs remains a significant hurdle because their large size (~150 kDa) makes mass spectrometry analysis extremely challenging.
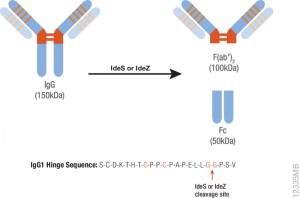
IdeS protease specifically cleaves IgGs into Fab and Fc fragments. This enzyme is highly specific and cleaves human IgG specifically at one site in the lower hinge region. Because of the exquisite specificity of the enzyme, it produces highly homogeneous Fc and Fab fragments which are then readily analyzed using techniques such as mass spectrometry or HPLC.
Are you looking for proteases to use in your research?
Explore our portfolio of proteases today.
One of the drawbacks of IdeS is that it exhibits poor activity against mouse IgGs. IdeZ Protease is an immunoglobulin-degrading enzyme from Streptococcus equi subspecies zooepidemicus. It is an engineered recombinant protease overexpressed in E. coli. Like IdeS Protease, IdeZ Protease specifically cleaves IgG molecules below the hinge region to yield F(ab′)2 and Fc fragments. Reduction of the digestion products produces three fragments of ~25kDa that are readily analyzed by LC-MS.
One of the key advantages of the IdeZ Protease is that it has significantly improved activity against mouse IgG2a and IgG3 subclasses compared to IdeS Protease. IdeZ Protease does not cleave mouse IgG1 or IgG2b.
Key technical parameters when digesting mouse IgGs utilizing IdeZ are the following:
• Add 1 unit of IdeZ Protease per 1µg of IgG to be digested.
• IdeZ Protease is most active in buffers at or near neutral pH. The recommended digestion buffer is 50mM sodium phosphate, 150mM NaCl (pH 6.6).
• Mouse IgG2a and IgG3 typically require 2–4 hours at 37°C for complete digestion.
• IdeZ Protease has a histidine tag for easy removal if so desired.
