
I confess that I struggled through biophysics, and my Bertil Hille textbook Ion Channels of Excitable Membranes lies neglected somewhere in a box in my basement (I have not tossed it into the recycle bin—I can’t bear too, I spent too much time bonding with that book in graduate school).
My struggles in that graduate class and my attendance at the seminars of my grad school colleagues who were conducting electrophysiological studies left me with a sincere awe and appreciation of both the genius and the artistry required to produce nice electrophysiology data. The people who are good at these experiments are artists—they have the golden touch when it comes to generating that megaohm seal between a piece of cell membrane and a finely pulled glass pipette. And, they are brilliant scientists, they really understand the physics, the chemistry and the biology of the cells they study from a perspective that very few scientists ever develop.
Electrophysiology data, which often demonstrate the gating of a single channel protein in response to a single stimulus in real time–ions crossing a membrane through a single protein–are amazing for their ability, unlike virtually any other experimental data for the story they can tell about what is going on in a cell in real time under physiological conditions.
So when I read the paper recently published by Mashuo et al. in Science Signaling “Distinct profiles of functional discrimination among G proteins determine the action of G protein-coupled receptors”, this sentence really caught my attention:
When constructs were ectopically expressed in HEK 293T/17 cells, we obtained very similar kinetics for the GPCR-driven responses between NanoBRET™ biosensors and the patch clamp recordings.
They continue:
Indeed, the activation rates that we observed were very similar to those of GPCR-stimulated GIRKs [G protein-coupled, inwardly rectifying K+ channel] in native cells, suggesting that the conditions of this assay closely match the in vivo setting. This finding further demonstrates the ability of the system to resolve the fast, physiological relevant kinetics of GPCR signaling.
A reporter biosensor that can resolve events similarly to patch clamping?! Amazing.
Those results have huge implications for improving our understanding many events as they occur in actual, physiological settings. Even in this one study, the sensitivity of the NanoBRET biosensor allowed the researchers to detect interactions between GPCRs and specific classes of G proteins that were beyond the resolution of previous attempts.
What they wanted to know
In this study, Mashuo and colleagues were attempting to better understand the diverse functional effects that G protein-coupled receptors (GPCRs) have on cell physiology. GPCRs are targets for 30–40% of the pharmaceuticals currently on the market. Even so, there is a great deal that remains unknown about how a specific GPCR generates a specific cellular response, and those unknowns mean pharmaceuticals can have unexpected off-target effects or be subject to cells developing resistance.
We do know for instance that interaction with specific ligands can determine whether a receptor interacts with the ß-arrestin scaffold or the G-proteins, and that changes the resulting downstream signaling cascade. There is also some evidence that the specific interactions of GPCRs with each other can affect precise effectors that are involved downstream signaling events.
Mashuo and colleagues proposed that additional specificity of downstream signaling events might be driven by the specificity and nature of the interaction of GPCRs with G-proteins, specifically the Gα subunits.
What they did
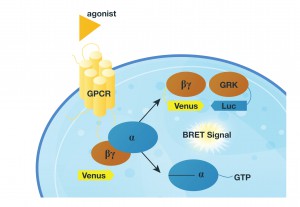 To test their proposal they designed a system in which they monitored the release of the Gßγ subunit after inducing GPCR activity with various agonists. They did this by monitoring the interaction Venus-tagged Gßγ subunit with a reporter (a luciferase-tagged effector-like peptide derived from G protein-coupled receptor kinase 3) in a BRET assay. If the GPCR interacts with the G-protein the Gßγ subunit is released and interacts with the reporter protein, and luminescence is produced (see figure).
To test their proposal they designed a system in which they monitored the release of the Gßγ subunit after inducing GPCR activity with various agonists. They did this by monitoring the interaction Venus-tagged Gßγ subunit with a reporter (a luciferase-tagged effector-like peptide derived from G protein-coupled receptor kinase 3) in a BRET assay. If the GPCR interacts with the G-protein the Gßγ subunit is released and interacts with the reporter protein, and luminescence is produced (see figure).
In their first attempts, they tagged the reporter peptide with Renilla luciferase, but they were unable to measure low-efficiency reactions and could not get good temporal resolution. So, they changed the reporter to NanoLuc® Luciferase, for the NanoBRET assay. Luminescent signal was considerably greater using the new luciferase, making it possible to record interaction kinetics with millisecond resolution.
What they learned, in brief
The authors of this study were able to detect signaling through Gα12/13 proteins, which had previously proved intractable, and when they looked at a wide variety of GPCR models, they were able to determine GPCRs produce different “fingerprints” of interactions with types of Gα subunits based on reaction time. They generated a host of GPCR fingerprints, showing for many receptors that the kinetics of G protein activation do not always correlate with the maximal amplitude of activation. Basically—who the G protein partner is and at when the interaction occurs are both questions that need to be answered to generate an accurate G-protein interaction “fingerprint” for a GPCR.
Additional Reading
Here’s the Paper:
Mashuo, I. et al. (2015) Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci. Signal. 8, ra123.
More about NanoBRET™ Assay
